01 [로지스틱 회귀 분석] 특징 데이터로 유방암 진단하기
사이킷런 의 유방암 진단 데이터셋 사용하기
import numpy as np
import pandas as pd
from sklearn.datasets import load_breast_cancer
b_cancer = load_breast_cancer()
print(b_cancer.DESCR)
.. _breast_cancer_dataset:
Breast cancer wisconsin (diagnostic) dataset
--------------------------------------------
**Data Set Characteristics:**
:Number of Instances: 569
:Number of Attributes: 30 numeric, predictive attributes and the class
:Attribute Information:
- radius (mean of distances from center to points on the perimeter)
- texture (standard deviation of gray-scale values)
- perimeter
- area
- smoothness (local variation in radius lengths)
- compactness (perimeter^2 / area - 1.0)
- concavity (severity of concave portions of the contour)
- concave points (number of concave portions of the contour)
- symmetry
- fractal dimension ("coastline approximation" - 1)
The mean, standard error, and "worst" or largest (mean of the three
worst/largest values) of these features were computed for each image,
resulting in 30 features. For instance, field 0 is Mean Radius, field
10 is Radius SE, field 20 is Worst Radius.
- class:
- WDBC-Malignant
- WDBC-Benign
:Summary Statistics:
===================================== ====== ======
Min Max
===================================== ====== ======
radius (mean): 6.981 28.11
texture (mean): 9.71 39.28
perimeter (mean): 43.79 188.5
area (mean): 143.5 2501.0
smoothness (mean): 0.053 0.163
compactness (mean): 0.019 0.345
concavity (mean): 0.0 0.427
concave points (mean): 0.0 0.201
symmetry (mean): 0.106 0.304
fractal dimension (mean): 0.05 0.097
radius (standard error): 0.112 2.873
texture (standard error): 0.36 4.885
perimeter (standard error): 0.757 21.98
area (standard error): 6.802 542.2
smoothness (standard error): 0.002 0.031
compactness (standard error): 0.002 0.135
concavity (standard error): 0.0 0.396
concave points (standard error): 0.0 0.053
symmetry (standard error): 0.008 0.079
fractal dimension (standard error): 0.001 0.03
radius (worst): 7.93 36.04
texture (worst): 12.02 49.54
perimeter (worst): 50.41 251.2
area (worst): 185.2 4254.0
smoothness (worst): 0.071 0.223
compactness (worst): 0.027 1.058
concavity (worst): 0.0 1.252
concave points (worst): 0.0 0.291
symmetry (worst): 0.156 0.664
fractal dimension (worst): 0.055 0.208
===================================== ====== ======
:Missing Attribute Values: None
:Class Distribution: 212 - Malignant, 357 - Benign
:Creator: Dr. William H. Wolberg, W. Nick Street, Olvi L. Mangasarian
:Donor: Nick Street
:Date: November, 1995
This is a copy of UCI ML Breast Cancer Wisconsin (Diagnostic) datasets.
https://goo.gl/U2Uwz2
Features are computed from a digitized image of a fine needle
aspirate (FNA) of a breast mass. They describe
characteristics of the cell nuclei present in the image.
Separating plane described above was obtained using
Multisurface Method-Tree (MSM-T) [K. P. Bennett, "Decision Tree
Construction Via Linear Programming." Proceedings of the 4th
Midwest Artificial Intelligence and Cognitive Science Society,
pp. 97-101, 1992], a classification method which uses linear
programming to construct a decision tree. Relevant features
were selected using an exhaustive search in the space of 1-4
features and 1-3 separating planes.
The actual linear program used to obtain the separating plane
in the 3-dimensional space is that described in:
[K. P. Bennett and O. L. Mangasarian: "Robust Linear
Programming Discrimination of Two Linearly Inseparable Sets",
Optimization Methods and Software 1, 1992, 23-34].
This database is also available through the UW CS ftp server:
ftp ftp.cs.wisc.edu
cd math-prog/cpo-dataset/machine-learn/WDBC/
.. topic:: References
- W.N. Street, W.H. Wolberg and O.L. Mangasarian. Nuclear feature extraction
for breast tumor diagnosis. IS&T/SPIE 1993 International Symposium on
Electronic Imaging: Science and Technology, volume 1905, pages 861-870,
San Jose, CA, 1993.
- O.L. Mangasarian, W.N. Street and W.H. Wolberg. Breast cancer diagnosis and
prognosis via linear programming. Operations Research, 43(4), pages 570-577,
July-August 1995.
- W.H. Wolberg, W.N. Street, and O.L. Mangasarian. Machine learning techniques
to diagnose breast cancer from fine-needle aspirates. Cancer Letters 77 (1994)
163-171.b_cancer_df = pd.DataFrame(b_cancer.data, columns = b_cancer.feature_names)
b_cancer_df['diagnosis'] = b_cancer.target
b_cancer_df.head()
데이터셋의 크기와 독립 변수 X가 되는 피처에 대한 정보
print('유방암 진단 데이터셋 크기: ', b_cancer_df.shape)
유방암 진단 데이터셋 크기: (569, 31)
b_cancer_df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 569 entries, 0 to 568
Data columns (total 31 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 mean radius 569 non-null float64
1 mean texture 569 non-null float64
2 mean perimeter 569 non-null float64
3 mean area 569 non-null float64
4 mean smoothness 569 non-null float64
5 mean compactness 569 non-null float64
6 mean concavity 569 non-null float64
7 mean concave points 569 non-null float64
8 mean symmetry 569 non-null float64
9 mean fractal dimension 569 non-null float64
10 radius error 569 non-null float64
11 texture error 569 non-null float64
12 perimeter error 569 non-null float64
13 area error 569 non-null float64
14 smoothness error 569 non-null float64
15 compactness error 569 non-null float64
16 concavity error 569 non-null float64
17 concave points error 569 non-null float64
18 symmetry error 569 non-null float64
19 fractal dimension error 569 non-null float64
20 worst radius 569 non-null float64
21 worst texture 569 non-null float64
22 worst perimeter 569 non-null float64
23 worst area 569 non-null float64
24 worst smoothness 569 non-null float64
25 worst compactness 569 non-null float64
26 worst concavity 569 non-null float64
27 worst concave points 569 non-null float64
28 worst symmetry 569 non-null float64
29 worst fractal dimension 569 non-null float64
30 diagnosis 569 non-null int64
dtypes: float64(30), int64(1)
memory usage: 137.9 KB로지스틱 회귀 분석에 피처로 사용할 데이터를 평균이 0, 분산이 1이 되는 정규 분포 형태로 맞춘다.
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
b_cancer_scaled = scaler.fit_transform(b_cancer.data)
print(b_cancer.data[0])
[1.799e+01 1.038e+01 1.228e+02 1.001e+03 1.184e-01 2.776e-01 3.001e-01
1.471e-01 2.419e-01 7.871e-02 1.095e+00 9.053e-01 8.589e+00 1.534e+02
6.399e-03 4.904e-02 5.373e-02 1.587e-02 3.003e-02 6.193e-03 2.538e+01
1.733e+01 1.846e+02 2.019e+03 1.622e-01 6.656e-01 7.119e-01 2.654e-01
4.601e-01 1.189e-01]
print(b_cancer_scaled[0])
[ 1.09706398 -2.07333501 1.26993369 0.9843749 1.56846633 3.28351467
2.65287398 2.53247522 2.21751501 2.25574689 2.48973393 -0.56526506
2.83303087 2.48757756 -0.21400165 1.31686157 0.72402616 0.66081994
1.14875667 0.90708308 1.88668963 -1.35929347 2.30360062 2.00123749
1.30768627 2.61666502 2.10952635 2.29607613 2.75062224 1.93701461]분석 모델 구축 및 결과 분석
로지스틱 회귀를 이용하여 분석 모델 구축하기
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
#X, Y 설정하기
Y = b_cancer_df['diagnosis']
X = b_cancer_scaled
#훈련용 데이터와 평가용 데이터 분할하기
X_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size = 0.3, random_state = 0)
#로지스틱 회귀 분석: (1) 모델 생성
lr_b_cancer = LogisticRegression()
#로지스틱 회귀 분석: (2) 모델 훈련
lr_b_cancer.fit(X_train, Y_train)
LogisticRegression()
#로지스틱 회귀 분석: (3) 평가 데이터에 대한 예측 수행 -> 예측 결과 Y_predict 구하기
Y_predict = lr_b_cancer.predict(X_test)생성한 모델의 성능 확인하기
from sklearn.metrics import confusion_matrix, accuracy_score
from sklearn.metrics import precision_score, recall_score, f1_score, roc_auc_score
#오차 행렬
confusion_matrix(Y_test, Y_predict)
array([[ 60, 3],
[ 1, 107]])
accuracy = accuracy_score(Y_test, Y_predict)
precision = precision_score(Y_test, Y_predict)
recall = recall_score(Y_test, Y_predict)
f1 = f1_score(Y_test, Y_predict)
roc_auc = roc_auc_score(Y_test, Y_predict)
print('정확도: {0:.3f}, 정밀도: {1:.3f}, 재현율: {2:.3f}, F1: {3:.3f}'.format(accuracy,precision,recall,f1))
정확도: 0.977, 정밀도: 0.973, 재현율: 0.991, F1: 0.982
print('ROC_AUC: {0:.3f}'.format(roc_auc))
ROC_AUC: 0.97202 [결정 트리 분석 + 산점도/선형 회귀 그래프] 센서 데이터로 움직임 분류하기
데이터 탐색
훈련용과 테스트용 데이터셋 확인하기
import numpy as np
import pandas as pd
pd.__version__
1.3.5
#피처 이름 파일 읽어오기
feature_name_df = pd.read_csv('/features.txt', sep = '\s+', header = None, names = ['index', 'feature_name'], engine = 'python')
feature_name_df.head()
feature_name_df.shape
(561, 2)
#index 제거하고, feature_name만 리스트로 저장
feature_name = feature_name_df.iloc[:, 1].values.tolist()
feature_name[:5]
['tBodyAcc-mean()-X',
'tBodyAcc-mean()-Y',
'tBodyAcc-mean()-Z',
'tBodyAcc-std()-X',
'tBodyAcc-std()-Y']
X_train = pd.read_csv('/X_train.txt', delim_whitespace=True, header=None, encoding='latin-1')
X_train.columns = feature_name
X_test = pd.read_csv('/X_test.txt', delim_whitespace=True, header=None, encoding='latin-1')
X_test.columns = feature_name
Y_train = pd.read_csv('/Y_train.txt', sep='\s+', header = None, names = ['action'], engine = 'python')
Y_test = pd.read_csv('/Y_test.txt', sep='\s+', header = None, names = ['action'], engine = 'python')
X_train.shape, Y_train.shape, X_test.shape, Y_test.shape
((7352, 561), (7352, 1), (2947, 561), (2947, 1))
X_train.head()
print(Y_train['action'].value_counts())
6 1407
5 1374
4 1286
1 1226
2 1073
3 986
Name: action, dtype: int64
label_name_df = pd.read_csv('/activity_labels.txt', sep = '\s+', header = None, names = ['index', 'label'], engine = 'python')
#index 제거하고, feature_name만 리스트로 저장
label_name = label_name_df.iloc[:, 1].values.tolist()
label_name
['WALKING',
'WALKING_UPSTAIRS',
'WALKING_DOWNSTAIRS',
'SITTING',
'STANDING',
'LAYING']분석 모델 구축 및 결과 분석
결정 트리 분류 분석 모델 구축하기
from sklearn.tree import DecisionTreeClassifier
#결정 트리 분류 분석: 모델 생성
dt_HAR = DecisionTreeClassifier(random_state=156)
#결정 트리 분류 분석: 모델 훈련
dt_HAR.fit(X_train, Y_train)
DecisionTreeClassifier(random_state=156)
#결정 트리 분류 분석: 평가 데이터에 예측 수행 -> 예측 결과로 Y_predict 구하기
Y_predict = dt_HAR.predict(X_test)생성한 모델의 성능 확인하고, 분류 정확도 높이기
from sklearn.metrics import accuracy_score
accuracy = accuracy_score(Y_test, Y_predict)
print('결정 트리 예측 정확도: {0:.4f}'.format(accuracy))
결정 트리 예측 정확도: 0.8548
print('결정 트리의 현재 하이퍼 매개변수: \n', dt_HAR.get_params())
결정 트리의 현재 하이퍼 매개변수:
{'ccp_alpha': 0.0, 'class_weight': None, 'criterion': 'gini', 'max_depth': None, 'max_features': None, 'max_leaf_nodes': None, 'min_impurity_decrease': 0.0, 'min_samples_leaf': 1, 'min_samples_split': 2, 'min_weight_fraction_leaf': 0.0, 'random_state': 156, 'splitter': 'best'}정확도를 검사하여 최적의 하이퍼 매개변수를 찾는 작업을 해주는 GridSearchCV 모듈을 사용해 보자
from sklearn.model_selection import GridSearchCV
params = {
'max_depth' : [6, 8, 10, 12, 16, 20, 24]
}
grid_cv = GridSearchCV(dt_HAR, param_grid = params, scoring = 'accuracy', cv = 5, return_train_score = True)
grid_cv.fit(X_train, Y_train)
GridSearchCV(cv=5, estimator=DecisionTreeClassifier(random_state=156),
param_grid={'max_depth': [6, 8, 10, 12, 16, 20, 24]},
return_train_score=True, scoring='accuracy')
cv_results_df = pd.DataFrame(grid_cv.cv_results_)
cv_results_df[['param_max_depth', 'mean_test_score', 'mean_train_score']]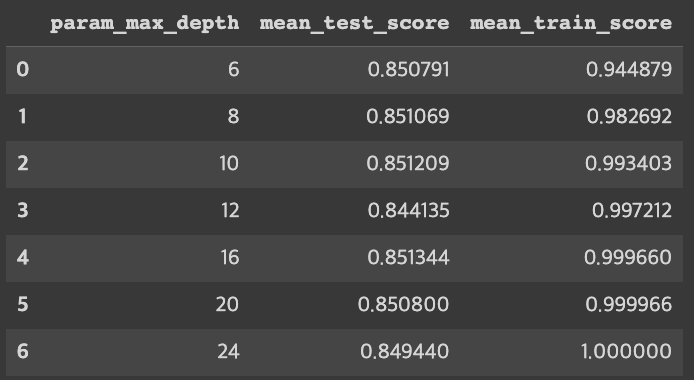
print('최고 평균 정확도: {0:.4f}, 최적 하이퍼 매개변수: {1}'.format(grid_cv.best_score_, grid_cv.best_params_))
최고 평균 정확도: 0.8513, 최적 하이퍼 매개변수: {'max_depth': 16}max_depth와 함께 min_sample_split을 조정하면서 최고의 평균 정확도 확인해 보자
params = {
'max_depth' : [8, 16, 20],
'min_samples_split' : [8, 16, 24]
}
grid_cv = GridSearchCV(dt_HAR, param_grid = params, scoring = 'accuracy', cv = 5, return_train_score = True)
grid_cv.fit(X_train, Y_train)
GridSearchCV(cv=5, estimator=DecisionTreeClassifier(random_state=156),
param_grid={'max_depth': [8, 16, 20],
'min_samples_split': [8, 16, 24]},
return_train_score=True, scoring='accuracy')
cv_results_df = pd.DataFrame(grid_cv.cv_results_)
cv_results_df[['param_max_depth', 'param_min_samples_split', 'mean_test_score', 'mean_train_score']]
print('최고 평균 정확도: {0:.4f}, 최적 하이퍼 매개변수: {1}'.format(grid_cv.best_score_, grid_cv.best_params_))
최고 평균 정확도: 0.8549, 최적 하이퍼 매개변수: {'max_depth': 8, 'min_samples_split': 16}최적모델(grid_cv.best_estimator_)을 사용하여 테스트 데이터에 대한 예측을 수행해 보자.
best_dt_HAR = grid_cv.best_estimator_
best_Y_predict = best_dt_HAR.predict(X_test)
best_accuracy = accuracy_score(Y_test, best_Y_predict)
print('best 결정 트리 예측 정확도: {0:.4f}'.format(best_accuracy))
best 결정 트리 예측 정확도: 0.8717결정 트리 모델은 feature_importances_ 속성을 사용하여 중요도가 높은 10개 피처를 찾아 그래프로 나타내보자.
import seaborn as sns
import matplotlib.pyplot as plt
feature_importance_values = best_dt_HAR.feature_importances_
feature_importance_values_s = pd.Series(feature_importance_values, index = X_train.columns)
feature_top10 = feature_importance_values_s.sort_values(ascending = False)[:10]
plt.figure(figsize = (10, 5))
plt.title('Feature Top 10')
sns.barplot(x = feature_top10, y = feature_top10.index)
plt.show()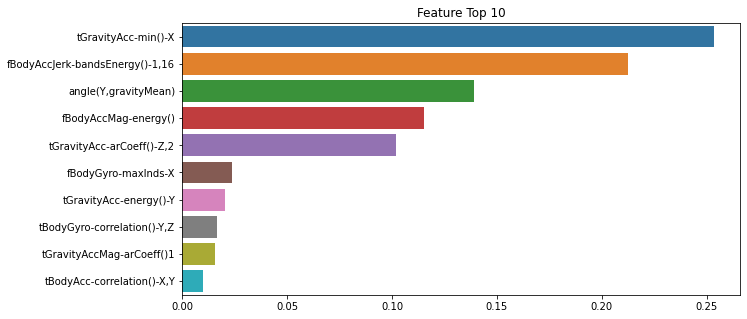
결과 시각화
결정 트리 모델의 트리 구조를 그림으로 시각화하기
!pip install graphviz
Looking in indexes: https://pypi.org/simple, https://us-python.pkg.dev/colab-wheels/public/simple/
Requirement already satisfied: graphviz in /usr/local/lib/python3.8/dist-packages (0.10.1)
from sklearn.tree import export_graphviz
#export_graphviz()의 호출 결과로 out_file로 지정된 tree.dot 파일 생성 export_graphviz(best_dt_HAR, out_file = "tree.dot", class_names = label_name, feature_names = feature_name, impurity = True, filled = Ture)
import graphviz
#위에서 생성된 tree.dot 파일을 Graphviz가 읽어서 시각화
with open("/tree.dot") as f:
dot_graph = f.read()
graphviz.Source(dot_graph)
'Python > 데이터 과학 기반의 파이썬 빅데이터 분석(한빛 아카데미)' 카테고리의 다른 글
| 데이터 과학 기반의 파이썬 빅데이터 분석 Chapter13 텍스트 마이닝 (2) | 2023.01.11 |
|---|---|
| 데이터 과학 기반의 파이썬 빅데이터 분석 Chapter12 군집분석 (0) | 2023.01.10 |
| 데이터 과학 기반의 파이썬 빅데이터 분석 Chapter10 회귀 분석 (0) | 2023.01.09 |
| 데이터 과학 기반의 파이썬 빅데이터 분석 Chapter09 지리 정보 분석 (0) | 2023.01.09 |
| 데이터 과학 기반의 파이썬 빅데이터 분석 Chapter08 텍스트 빈도 분석 (0) | 2023.01.08 |



